

Since 2008, VRVis has been developing complete platforms for web-based management of the visualisation, integration and mining of brain data. The USP of the system is the very high-performing spatial indexing methods, which permit intuitive and almost real-time browsing through the content of tens of thousands of 3D data sets for similar or overlapping substructures. Our developments are based on our modular Brain* framework.
Our systems are now installed in numerous institutions and offer (partially public) access to large collections of data. The BrainBase system is currently hosting collections of data on the larval and adult brain of Drosophila melanogaster and the zebrafish. (See for example larvalbrain.org, fruitfly.tefor.net, zebrafish.tefor.net).
Together with neuroscientists at the Haubensak Lab and Boehringer-Ingelheim, we are developing BrainTrawler for collections of data on the brain of mice and humans. Using spatial indexing, we are able to analyse gene expression data and complex brain networks in real time. Classifying the data to hierarchically organised anatomical structures make it possible to search at different anatomical levels. Together with intuitive network visualisation, iterative visual queries and quantitative information, BrainTrawler enables multimodal networks to be genetically dissected at local/global level within the spatial context.
Selection of publications
Project: 4D Larvalbrain
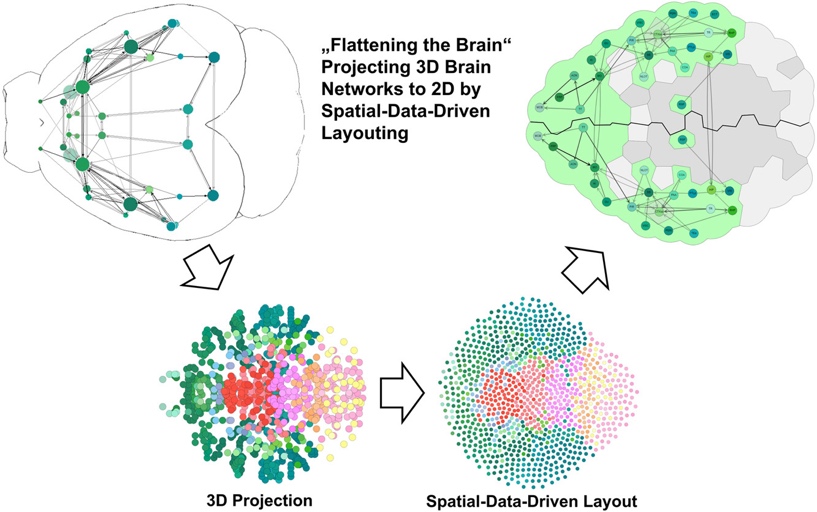
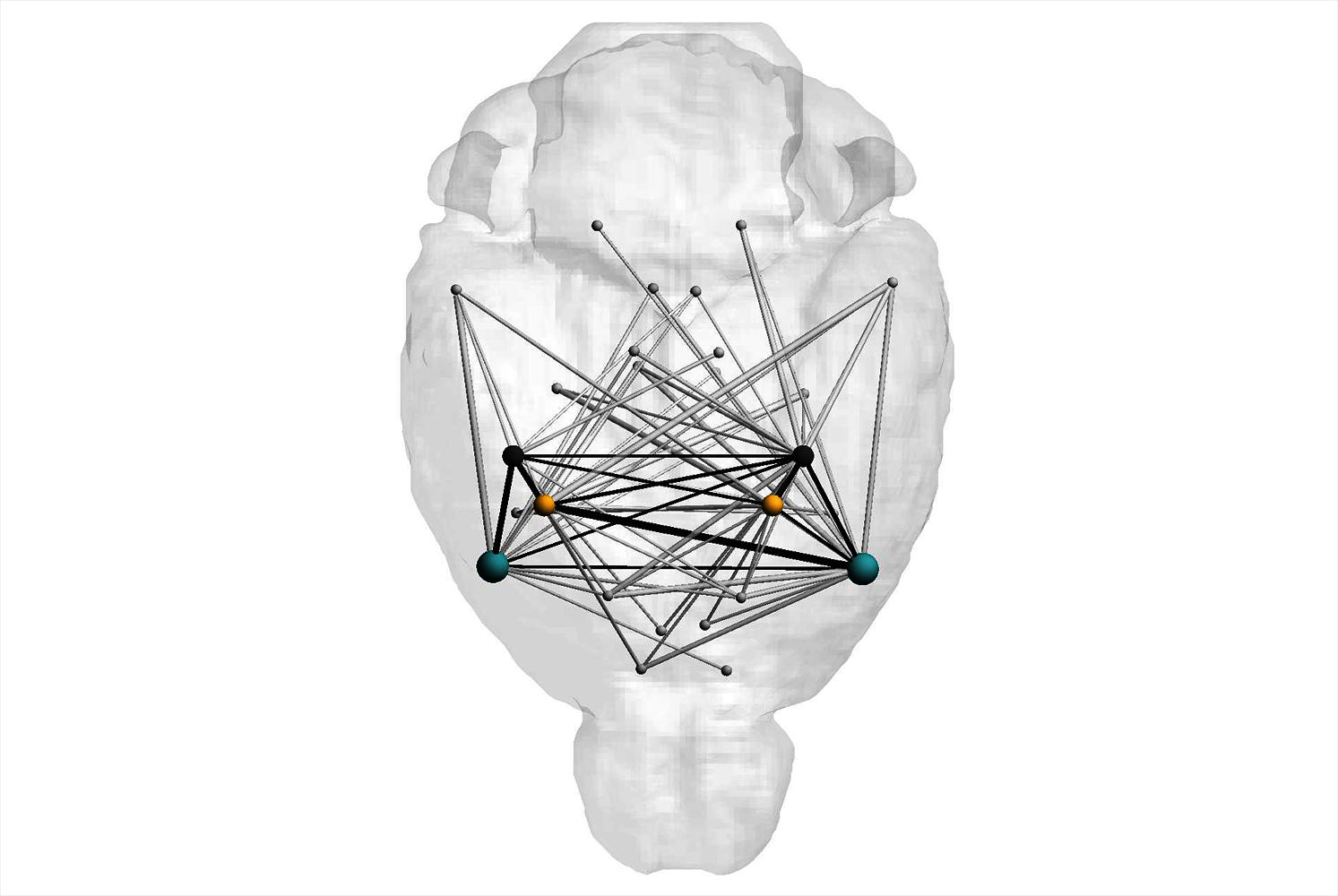
The visualisation of neuroscientific data for publications and also for interactive data exploration within the context of the Brain* system is an important research area of VRVis. For example, several publications focussed particularly on the visualisation and interactive analysis of brain network data, as well as the implementation of very high-performance web-based 2D and 3D visualisation of brain data (see for example larvalbrain.org).
We develop and explore tailor-made data science and visual analytics solutions to analyse the often very demanding, heterogeneous data from behavioural and pharmacological experiments. There is a wide variety of data types: In addition to recordings of neuronal activities, we also apply our methods to data which has been extracted from video recordings of behavioural studies.
In addition to “conventional” methods of machine and deep learning, such as classifications and cluster analyses, we also use methods from information theory or psychophysical models.
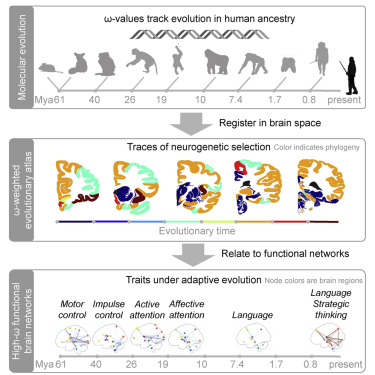
Computational Neuroscience gains new insights based on existing data and mathematical methods. In cooperation with various partners we developed new approaches to e.g. calculate new functional brain regions fusing tens of thousands of image data sets of neurons, or to reconstruct functional partial networks which correlate with certain genes. Currently we attempt to map the origin of human cognitive features by combining evolutionary genomics with functional neuroanatomy to recreate the neuro-genetical development of human brain functions more comprehensively and in a more direct way.
It is our goal to enhance the performance of complex calculations on extremely big data sets. For this purpose, we research and develop highly parallelized solutions and use selected available high-performance computing infrastructure.
References
The tools of Extended Reality – Augmented Reality (AR) and Virtual Reality (VR) – are being used more and more often in the Life Sciences. Digital Realities help enhance scientific experiments, trainings and procedures by creating cheap and controllable immersive environments true to reality.
For the department of Neurobiology at the University of Vienna VRVis created a setup for the research into the reactions of hunting spiders. The spider moves on a ball of about one meter in diameter inside a projected virtual environment. A camera picks up on the movements and the ball is rotated accordingly, so that the spider always remains in the center of the setup. In this manner, various visual stimulations can be produced in correlation with spider movements, even ones, that would not be possible in a purely physical setup.
References

Within the project Larvalbrain 2.0, a dynamic multi-scale multi-level atlas and data collection of structural, molecular, physiological, and behavioral results of Drosophila melanogaster larvae will be established.

For centuries, neuroscientists have been mapping the brain. Until now, the step from simple maps to a generally accepted model has proved to be extremely difficult. In this project, a 4D atlas of the brain of the fruit fly larva is being built.
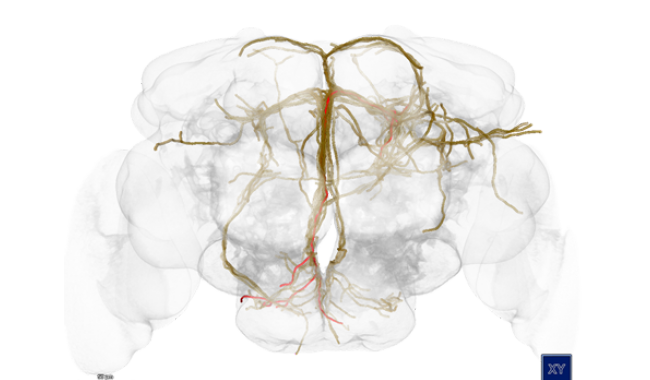
Understanding how the brain works is one of the biggest challenges addressed by neuroscientists today. Modern neuroscience research is extremely data-intensive and requires special software infrastructures to enable and accelerate the discovery of the complex interplay of genes, structure and function.

VRVis contributes data analytics and visualization tools tailored to support and accelerate research of the Haubensak Group at the Institute of Molecular Pathology Vienna.
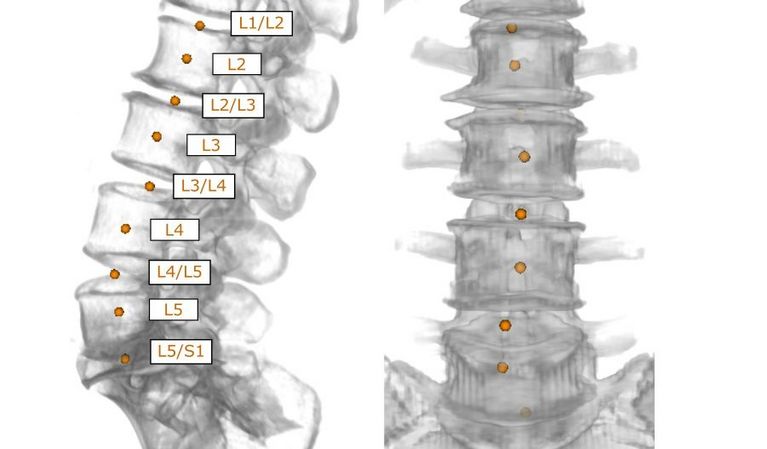
The long-term vision of this applied research project is to use available data resources to improve image-based diagnostics based on complex data in daily clinical routine.
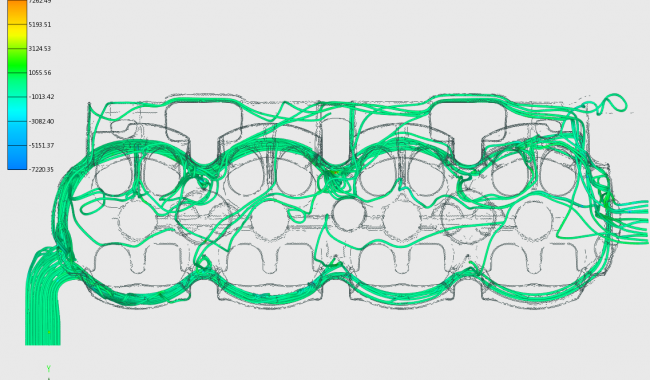
The strategic project forms the organizational and scientific hub for the realization of an area wide integrative visual computing approach. It covers joint strategic research and development on fundamental challenges in all application projects.
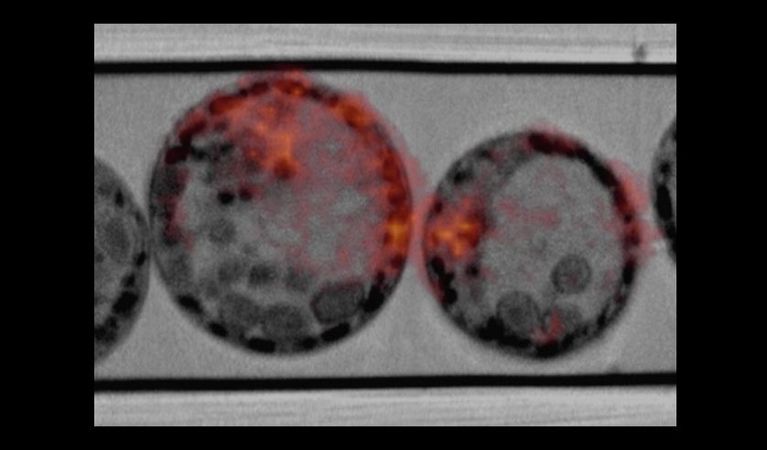
COMULIS is an EU-funded COST Action that aims at fueling collaborations in the field of correlated multimodal imaging (CMI).

VRVis a founding member of the Austrian BioImaging/CMI, which is a professional consortium of multiple Austrian science institutions and the official Austrian Euro-BioImaging initiative.

During the 40th anniversary celebration of the Austrian Computer Society (OCG) on June 9, 2015, Johannes Sorger was awarded this year's OCG sponsorship award.

The paper "Visual and Quantitative Analysis of Higher Order Arborization Overlaps for Neural Circuit Research" was awarded.

The web-based tool "neuroMAP" of VRVis and IMP received the Best Paper Award of the IEEE BioVis 2013.